Model Classes¶
Class Description¶
| Class Name | Class ID | Common Name | AphiaID | URN | Grouping | HAB | Description | Manual Classifier Notes | Example Images |
|---|---|---|---|---|---|---|---|---|---|
| Akashiwo | 0 | Akashiwo sanguinea | 232546 | urn:lsid:marinespecies.org:taxname:232546 | Dinoflagellate | Y | Monophyletic, marine dinoflagellate | large single cells, kidney-shaped when viewed from top or bottom | |
| Alexandrium_singlet | 1 | Alexandium sp. | 109470 | urn:lsid:marinespecies.org:taxname:109470 | Dinoflagellate | Y |
|
Originally class was broken out for singles, doublets, etc. But now all Alex is in this class, so the singlet is a misnomer | http://akashiwo.oceandatacenter.ucsc.edu:8000/image?image=00032&bin=D20161013T081022_IFCB104 |
| Amy_Gony_Protoc | 2 | Amylax, Gonyaulax or Protoceratium | 109428 | urn:lsid:marinespecies.org:taxname:109428 | Dinoflagellate | Y | Descision was made not to distinguish between these, originally with the RF. | “Feet” and points visible. | |
| Asterionellopsis | 3 | Asterionellopsis | 149138 | urn:lsid:marinespecies.org:taxname:149138 | Diatom | N | Common species include A. glacialis, A. kariana | ||
| Boreadinium | 4 | Boredadinium pisiforme | 110067 | urn:lsid:marinespecies.org:taxname:110067 | Dinoflagellate | N | Slightly compressed thecate cells | <45 um | |
| Centric | 5 | Centric Diatoms | 148965 | urn:lsid:marinespecies.org:taxname:148965 | Diatom | N | Order level. Round, or square/rectangle in side view | if longer than wide, put in det_cer_lau | |
| Ceratium | 6 | Ceratium | 109506 | urn:lsid:marinespecies.org:taxname:109506 | Dinoflagellate | N | All morphologies | Consider splitting up in future | |
| Chaetoceros | 7 | Chaetoceros | 148985 | urn:lsid:marinespecies.org:taxname:148985 | Diatom | N | Single and Chained | if setae are forked, Bacteriastrum | |
| Ciliates | 8 | Ciliates | 11 | urn:lsid:marinespecies.org:taxname:11 | Cilliate | N | Small Cilliates | ie not belonging to finer scoped classes | |
| Clusterflagellate | 9 | Clusteredflagellate | 7 | urn:lsid:marinespecies.org:taxname:7 | Other | N | Not well described, clusted flagellates | Posssibly Synura? | |
| Cochlodinium | 10 | Margalefidinium polykrikoides | 990876 | urn:lsid:marinespecies.org:taxname:990876 | Dinoflagellate | Y | Named was updated, but class name has not been updated. | ||
| Corethron | 11 | Corethron | 149109 | urn:lsid:marinespecies.org:taxname:149109 | Diatom | N | Diatom, Need to check if species can be resolve | ||
| Cryptophyte | 12 | Cryptophyte | 17638 | urn:lsid:marinespecies.org:taxname:17638 | Other | N | Biflagellate, with ejectsomes | Small teardrop shaped | |
| Cyl_Nitz | 13 | Nitzschia/Cylindrotheca | 149002 | urn:lsid:marinespecies.org:taxname:149002 | Diatom | N | Combindation of the two genera, Nitzschia and Cylindrotheca | Pennate with distinct rounded center + spines, not uniform thickness. | |
| Det_Cer_Lau | 14 | Detonula, Cerataulina, Lauderia | 148899 | urn:lsid:marinespecies.org:taxname:148899 | Diatom | N | Square or rectangular cells. Chain or single celled | If chains, cells must be touching. if single cells and wider than long, put in Centric | |
| Dictyocha | 15 | Dictocha | 157258 | urn:lsid:marinespecies.org:taxname:157258 | Other | N | Silicoflagellate | ||
| Dinophysis | 16 | Dinophysis | 19542 | urn:lsid:marinespecies.org:taxname:19542 | Dinoflagellate | Y | Morphologies are not currently disentangled | Rounded bottom and two “fins” | |
| Ditylum | 17 | Ditylum | 149022 | urn:lsid:marinespecies.org:taxname:149022 | Diatom | N | Large rolling-pin shaped diatom | Can be triangular in cross section. | |
| Entomoneis | 18 | Entomoneis | 156598 | urn:lsid:marinespecies.org:taxname:156598 | Diatom | N | Common in SF Bay | butterfly-shaped diatom, “fins” are transparent and sometimes out of frame | |
| Eucampia | 19 | Eucampia | 616809 | urn:lsid:marinespecies.org:taxname:616809 | Diatom | N | Ecuampia zoodiacus, bloom forming | ||
| FlagMix | 20 | Flagellates | 1 | urn:lsid:marinespecies.org:taxname:1 | Other | N | Catch all Flagellate, generally not enough resolution to | any flagellates except Pyramimonas | |
| Guin_Dact | 21 | Guinardia/Dactyliosolen | 149068 | urn:lsid:marinespecies.org:taxname:616811 | Diatom | N | Curved cells as opposed to Detonula, Cerataulina, Lauderia |
|
|
| Gymnodinium | 22 | Gymnodinium | 109475 | urn:lsid:marinespecies.org:taxname:109475 | Dinoflagellate | Y | Many species | ||
| Gyrodinium | 23 | Gyrodinium | 109476 | urn:lsid:marinespecies.org:taxname:109476 | Dinoflagellate | N | |||
| Hemiaulus | 24 | Hemiaulus | 163248 | urn:lsid:marinespecies.org:taxname:163248 | Diatom | N | Not to be confused with Eucampia. | Straight in valve view, curved in girdle view. Can have slightly convex ends, or straighter ends with horns | |
| Leptocylindrus | 25 | Leptocylindrus | 149038 | urn:lsid:marinespecies.org:taxname:149038 | Diatom | N | |||
| Licmophora | 26 | Licmophora | 149342 | urn:lsid:marinespecies.org:taxname:149342 | Diatom | N | |||
| Lingulodinium | 27 | Lingulodinium | 231799 | urn:lsid:marinespecies.org:taxname:231799 | Dinoflagellate | Y | |||
| Mesodinium | 28 | Mesodinium | 232069 | urn:lsid:marinespecies.org:taxname:232069 | Cilliate | N | Mesodinium rubrum. | Orientation can make this hard to distinguish from other cilliates. | |
| NanoP_less10 | 29 | Nano Plankton, < 10 μm | 1 | urn:lsid:marinespecies.org:taxname:1 | Other | N | Catch-all group for small (<10 um) cells that can’t be distinguished. | ||
| Odontella | 30 | Odontella | 148963 | urn:lsid:marinespecies.org:taxname:148963 | Diatom | N | |||
| Pennate | 31 | Pennate Diatom | 148899 | urn:lsid:marinespecies.org:taxname:148899 | Diatom | N | Single cell (not-chain) pennate diatoms | single or rafted pennate cells, includes single pseudos | |
| Peridinium | 32 | Peridinium | 109549 | urn:lsid:marinespecies.org:taxname:109549 | Dinoflagellate | N | Unarmored, dark dinoflagellate. No “feet” | Possibly Gymnodinium (unarmoured) but we’ve been putting them here. | |
| Phaeocystis | 33 | Phaeocystis | 115088 | urn:lsid:marinespecies.org:taxname:115088 | Other | N | clusters of small cells | ||
| Pleurosigma | 34 | Pleurosigma | 149181 | urn:lsid:marinespecies.org:taxname:149181 | Other | Y | |||
| Polykrikos | 35 | Polykrikos | 109485 | urn:lsid:marinespecies.org:taxname:109485 | Dinoflagellate | N | |||
| Prorocentrum | 36 | Prorocentrum | 109566 | urn:lsid:marinespecies.org:taxname:109566 | Dinoflagellate | Y | |||
| Protoperidinium | 37 | Protoperidinium | 109553 | urn:lsid:marinespecies.org:taxname:109553 | Dinoflagellate | Y | |||
| Pseudo-nitzschia | 38 | Pseudo-nitzschia | 149151 | urn:lsid:marinespecies.org:taxname:149151 | Diatom | Y | |||
| Pyramimonas | 39 | Pyramimonas | 134529 | urn:lsid:marinespecies.org:taxname:134529 | Other | N | |||
| Rhiz_Prob | 40 | Rhizosolenia/Proboscia | 149068 | urn:lsid:marinespecies.org:taxname:149068 | Diatom | N | |||
| Scrip_Het | 41 | Scrippsiella/Heterocapsa | 109394 | urn:lsid:marinespecies.org:taxname:109394 | Dinoflagellate | N | |||
| Skeletonema | 42 | Skeletonema | 149073 | urn:lsid:marinespecies.org:taxname:149073 | Diatom | N | |||
| Thalassionema | 43 | Thalassionema | 149092 | urn:lsid:marinespecies.org:taxname:149092 | Diatom | N | Pennate with squared ends, zig zag chains | Single cells go in Pennate | |
| Thalassiosira | 44 | Thalassiosira | 148912 | urn:lsid:marinespecies.org:taxname:148912 | Diatom | N | Two or more cells in a chain | Single cells go in Centric | |
| Tiarina | 45 | Tiarina | 247913 | urn:lsid:marinespecies.org:taxname:247913 | Other | N | |||
| Tintinnid | 46 | Tintinnid | 732976 | urn:lsid:marinespecies.org:taxname:732976 | Cilliate | N | |||
| Tontonia | 47 | Tontonia | 101196 | urn:lsid:marinespecies.org:taxname:101196 | Cilliate | N | |||
| Torodinium | 48 | Torodinium | 109479 | urn:lsid:marinespecies.org:taxname:109479 | Dinoflagellate | N | Large, blimp-shaped dino | ||
| Tropidoneis | 49 | Tropidoneis | 149518 | urn:lsid:marinespecies.org:taxname:149518 | Diatom | N | |||
| Vicicitus | 50 | Vicicitus | 707571 | urn:lsid:marinespecies.org:taxname:707571 | Diatom | N | Round, like a large centric, but lumpy | ||
| unclassified | 51 | Unclassified | urn:lsid:marinespecies.org:taxname: | Other | Classification lacked confidence to asssign a class |
Class-Specific Thresholds¶
Class-specific thresholds are an important processing step for accounting for bias in the training dataset. Nonuniformity in the training data reflect both practical desicions made by the creators of the data set and the natural distribution of observered phytoplankton classes. In plain terms, the result is that the model is more likely to guess that something belongs to the more commonly observed classes. In that case, we require a high threshold in that predication in order to assign confidence. Conversely when a less common class is observed, it makes sense to have a lower threshold to assign confidence.
Thresholds can be generated for each class by incrementally increasing a threshold from 0 to 1 and calculating the resulting metrics. In this case, the F-score was used to balance recall and percision. The threshold values where F-score begins to decrease (diff > 0.005) are the class-specific thresholds. Thresolds are calculated for each class and are shown in the table below.
Note
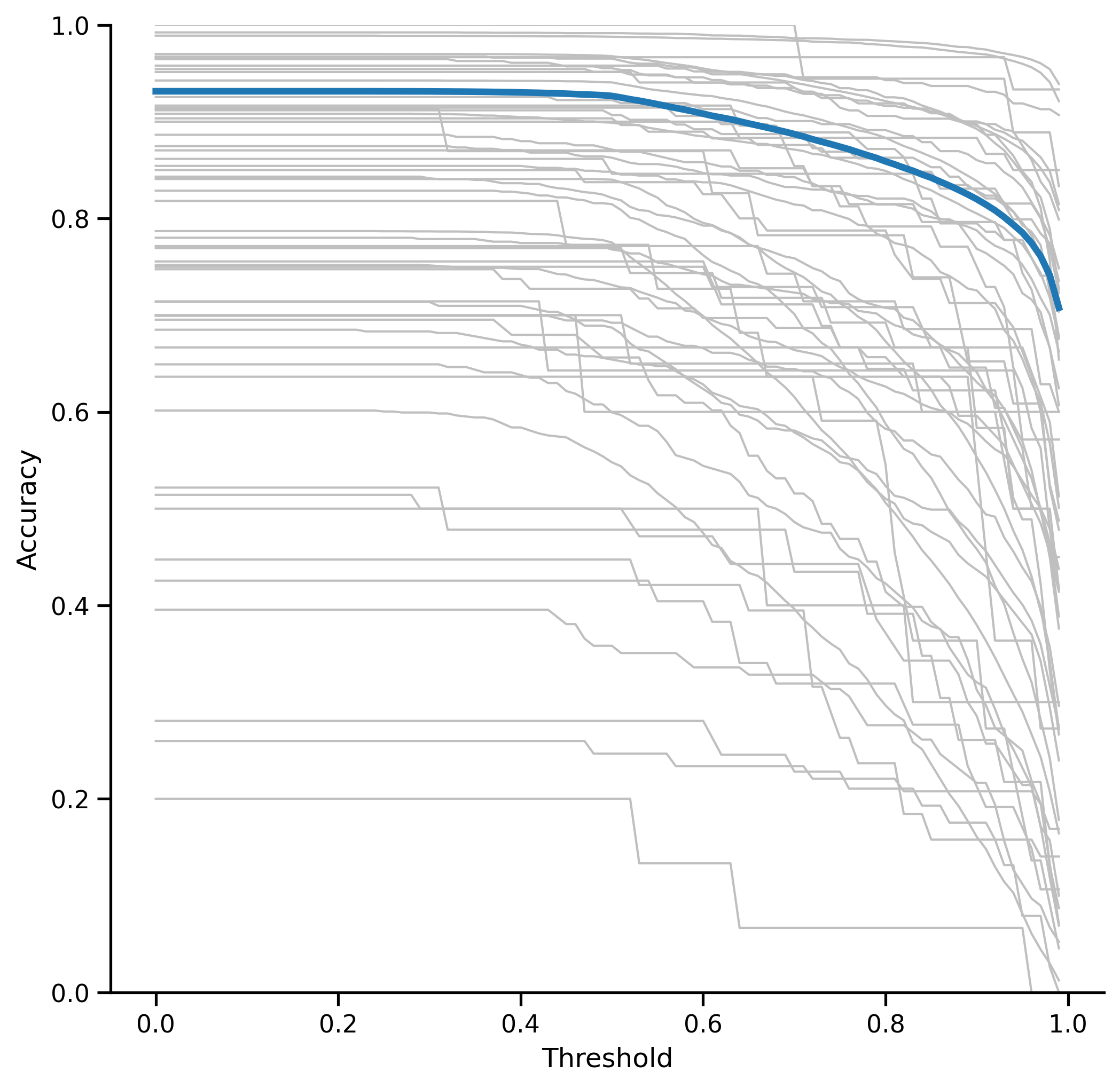
REPLACE WITH UP-TO-DATE Each curve shows the accuracy as the threshold is increased (up to .99). The point where the curve begins to decrease is the threshold value for that class.
| Class Name | Threshold |
|---|---|
| Akashiwo | 0.95 |
| Alexandrium_singlet | 0.49 |
| Amy_Gony_Protoc | 0.35 |
| Asterionellopsis | 0.56 |
| Boreadinium | 0.45 |
| Centric | 0.7 |
| Ceratium | 0.95 |
| Chaetoceros | 0.89 |
| Ciliates | 0.49 |
| Clusterflagellate | 0.55 |
| Cochlodinium | 0.48 |
| Corethron | 0.29 |
| Cryptophyte | 0.38 |
| Cyl_Nitz | 0.56 |
| Det_Cer_Lau | 0.52 |
| Dictyocha | 0.6 |
| Dinophysis | 0.92 |
| Ditylum | 0.27 |
| Entomoneis | 0.38 |
| Eucampia | 0.88 |
| FlagMix | 0.81 |
| Guin_Dact | 0.37 |
| Gymnodinium | 0.42 |
| Gyrodinium | 0.63 |
| Hemiaulus | 0.47 |
| Leptocylindrus | 0.62 |
| Licmophora | 0.57 |
| Lingulodinium | 0.56 |
| Mesodinium | 0.39 |
| NanoP_less10 | 0.92 |
| Odontella | 0.51 |
| Pennate | 0.81 |
| Peridinium | 0.44 |
| Phaeocystis | 0.81 |
| Pleurosigma | 0.35 |
| Polykrikos | 0.79 |
| Prorocentrum | 0.92 |
| Protoperidinium | 0.73 |
| Pseudo-nitzschia | 0.62 |
| Pyramimonas | 0.4 |
| Rhiz_Prob | 0.47 |
| Scrip_Het | 0.71 |
| Skeletonema | 0.43 |
| Thalassionema | 0.62 |
| Thalassiosira | 0.36 |
| Tiarina | 0.44 |
| Tintinnid | 0.94 |
| Tontonia | 0.46 |
| Torodinium | 0.49 |
| Tropidoneis | 0.45 |
| Vicicitus | 0.41 |